Machine learning model uncovers new drug design opportunities
× close
Pathogens are nothing if not adaptable, and their ability to protect themselves against antibiotics increasingly poses a public health concern. A research team led by Los Alamos National Laboratory has used machine learning, an application of artificial intelligence, to identify molecular properties that could guide the discovery of new types of antibiotics, especially among pathogens deemed critical by the World Health Organization due to their high bacterial resistance.
The findings are published in the journal Communications Chemistry.
“Some pathogens have properties that make them very effective at resisting antibiotics,” said Gnana Gnanakaran, scientist at Los Alamos. “The discovery of specific compounds able to permeate and inhibit some pathogens is a needle-in-the-haystack challenge due to the vast heterogeneity and depth of the chemical space, and the complexity of the molecular interactions across bacterial membranes. The approach we employ is capable of probing the bacterium-specific, molecular-level profiles necessary that can be built on for successful drug development.”
Bacterial defenses against antibiotics
Gram-negative bacteria have an outer membrane less permeable to being breached by compounds, such as those that make up antibiotics, and the bacteria can also expel compounds that happen to get inside, curtailing the effectiveness of an antibiotic.
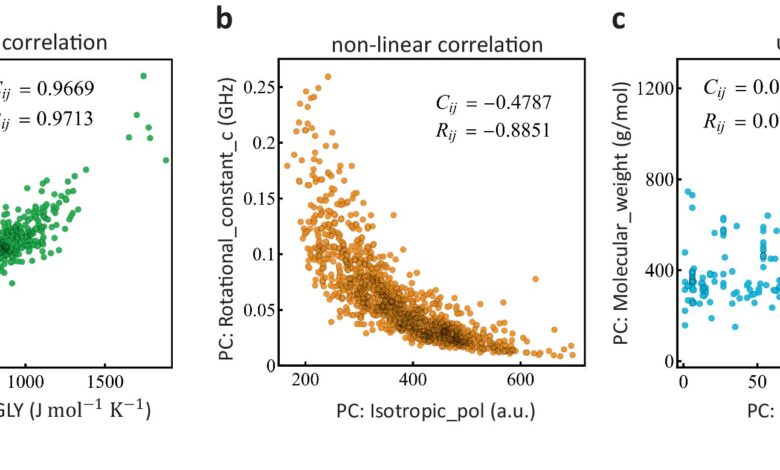
Data-driven models have potential to identify molecular properties that could overcome such bacterial defenses, but accurate calculations to make those determinations are challenging and use extensive computing resources. Chemically diverse compounds may contain many relevant properties; the machine learning-driven study reduced the relevant spectrum of those properties and established empirical rules that would predict the compound’s ability to permeate the bacteria’s outer membrane.
Machine learning model identifies pathogen-fighting properties
Focusing specifically on the gram-negative bacteria Pseudomonas aeruginosa, the research team developed a machine learning model to identify the relevant descriptors associated with compounds and predict those compounds’ success in permeating bacteria’s outer membranes and avoiding expulsion. The team relied on high-performance computing capabilities at Los Alamos to extract the molecular properties of permeation from simulations that considered 1,260 chemically diverse compounds as they traverse the bacterial membrane.
Their analysis sheds new light on the key properties drug candidates need to effectively permeate Pseudomonas aeruginosa, and opens the gate to similar data-driven studies in other gram-negative pathogens.
“The machine learning techniques we’ve employed in this analysis point to a promising approach for similar data-driven studies in other biological membranes, including the blood-brain barrier,” Gnanakaran said.
More information:
Pedro D. Manrique et al, Predicting permeation of compounds across the outer membrane of P. aeruginosa using molecular descriptors, Communications Chemistry (2024). DOI: 10.1038/s42004-024-01161-y
Journal information:
Communications Chemistry